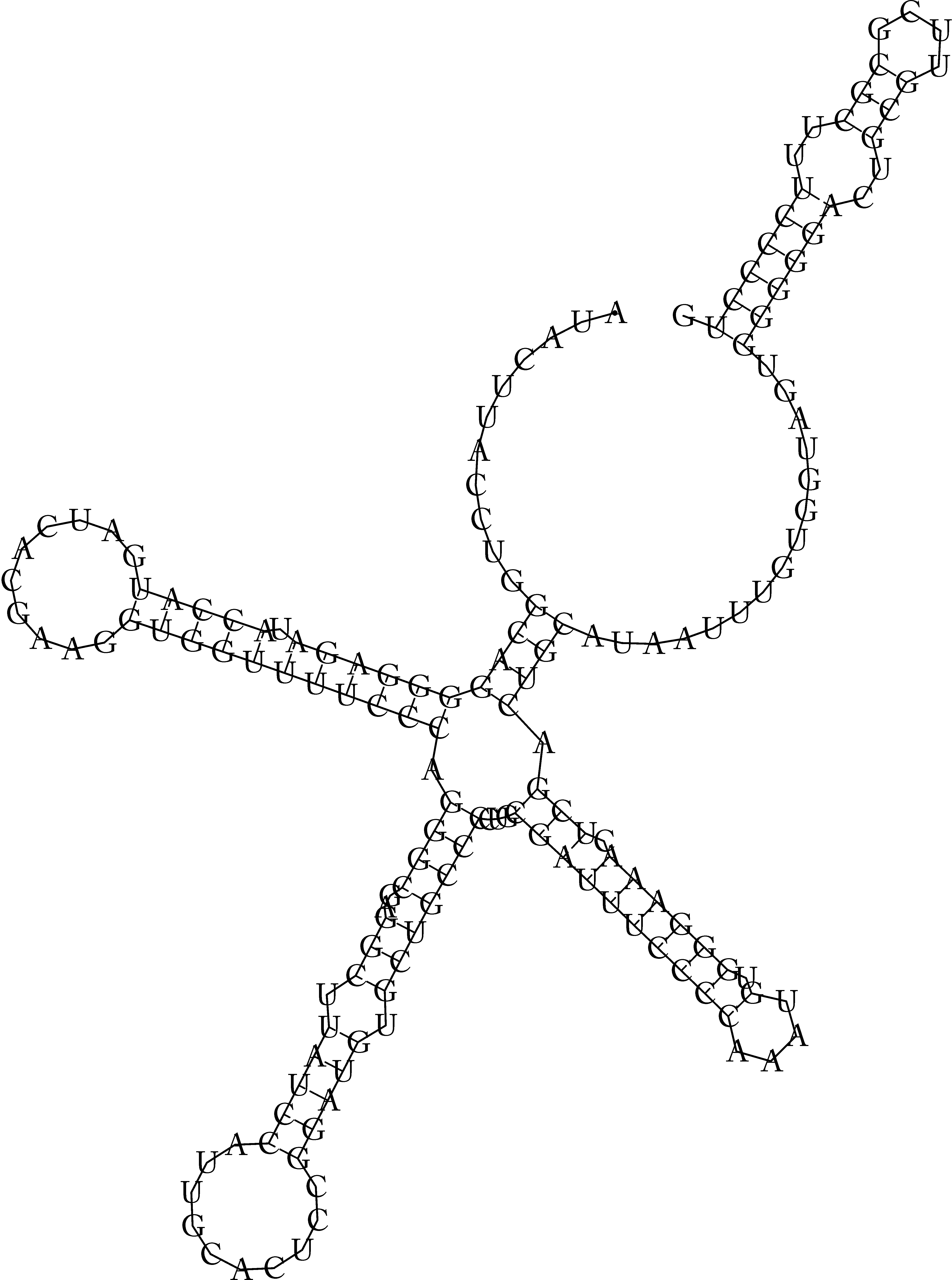3. Use cases¶
Here we provide three cases of the use of the cpPredictor server. They represent situations related to RNA secondary structure prediction that can not be worked out with available prediction methods, but can be worked out with cpPredictor through template-based prediction. The use cases are based on templates that are either related or unrelated to the RNAs under investigation. The use cases also demonstrate advanced usage of cpPredictor and its full capabilities.
3.1. Use case #1: Characterization of RNA sequences using a template structure¶
This use case demonstrates the situation, when we have a set of uncharacterized RNA sequences and a secondary structure, and want to characterize the sequences using the structure, i.e. to know if the sequences are able to adopt the structure and how well.
To that end, RNAs belonging to two different families, u1 and 6S RNAs are used as query RNAs. 5 sequences of each family are mixed together (see Text box 3.1)) and used as uncharacterized sequences.
Text box 3.1. Sequences of u1 and 6S RNAs, used as query sequences.
As a template, B. subtilis 6S RNA, a member of one of the families, is used (see Text box 3.2).
Text box 3.2. Secondary structure of B. subtilis 6S RNA used as a structural template.
The query sequences and the template structure are pasted into the appropriate text boxes on the submission page as shown in Figure 3.1. To get z-scores for the predicted structures, choose ‘Compute z-scores from 10 samples (quick)’ in the ‘Compute z-score’ option.
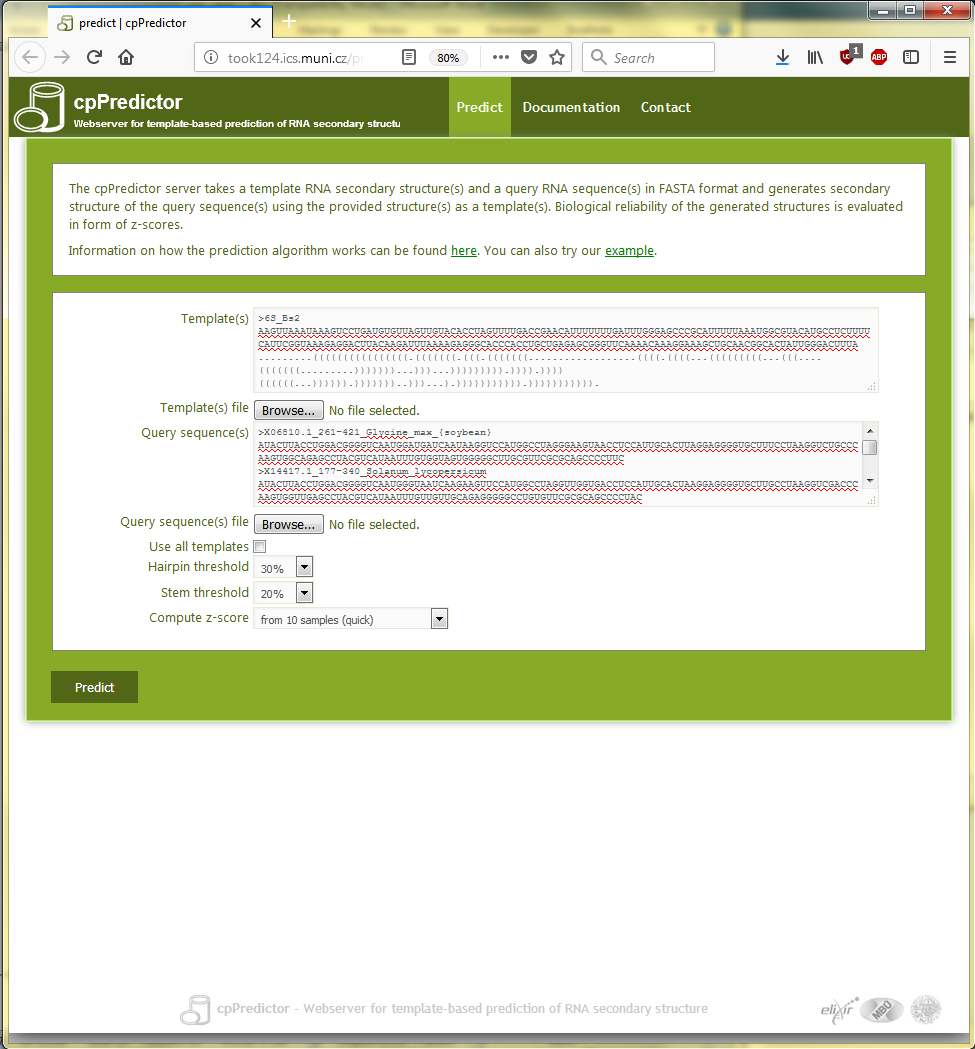
Figure 3.1. The submission page of cpPredictor web server filled up with query sequences and a template structure for the use case #1: Characterization of RNA sequences.
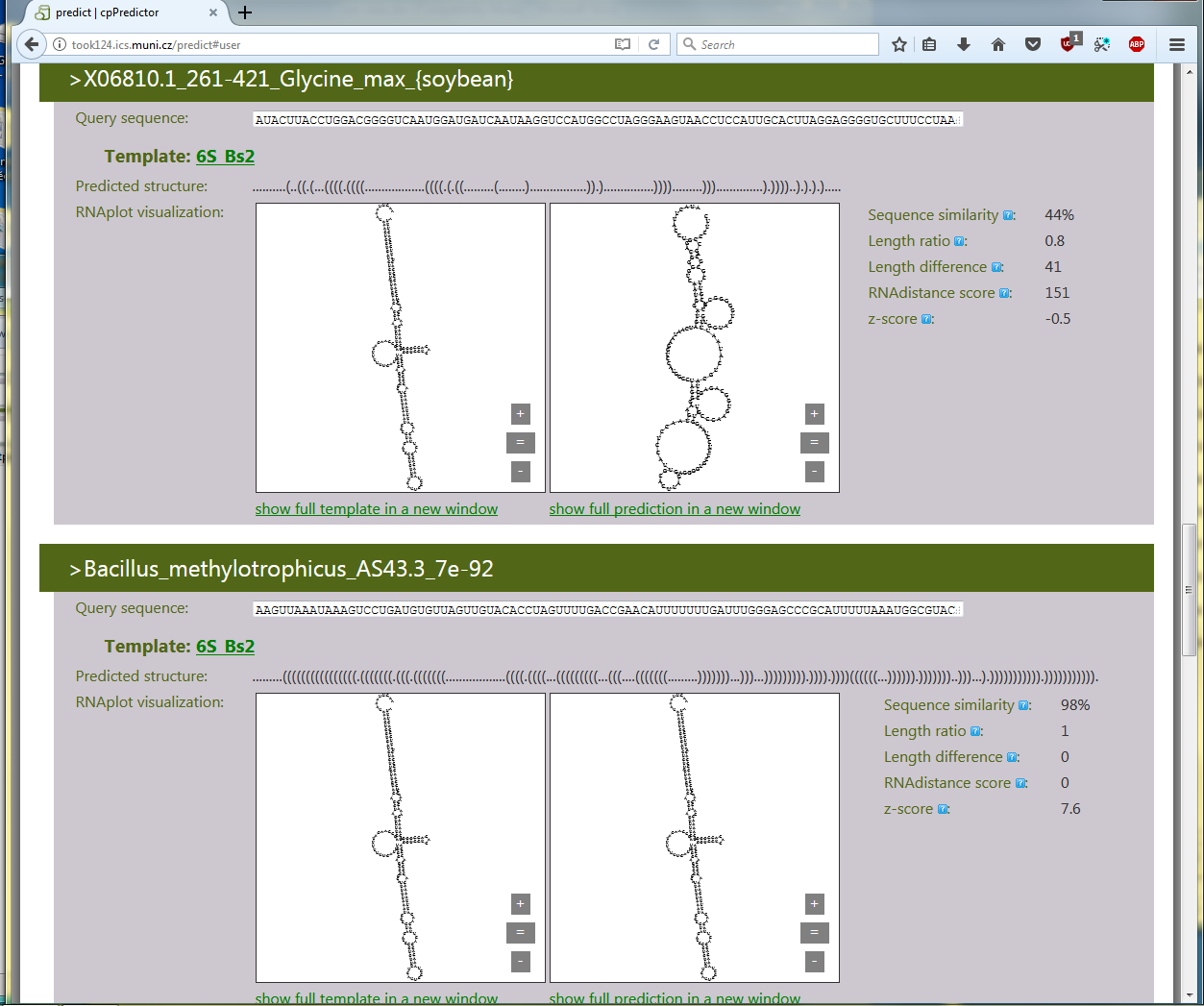
The cpPredictor output consists of generated structures together with their characteristics. Examples of two query RNAs copied from the complete cpPredictor output are shown in Figure 3.2. The first of them is Glycine max u1 RNA, the other Bacillus methylotrophicus 6S RNA. cpPredictor provides their secondary structures generated using the B. subtilis 6S RNA as a structural template. The upper secondary structure is unreliable, the lower is reliable. The reliability is indicated by z-scores of the predicted structures, and also can be recognized visually. The reliability shows that the sequence of the upper RNA is not able to adopt a proper 6S RNA structure and therefore is not 6S RNA. The other RNA is 6S RNA as it adopts a proper 6S RNA structure.
This way, query RNAs are characterized by both their secondary structures and the reliability of their secondary structures.
3.2. Use case #2: Characterization of RNA sequences including fragments using a template structure¶
This use case demonstrates the use of cpPredictor for characterization of RNA sequences including fragments using a template structure without knowing whether the template is or is not related to the characterized RNAs.
Sequences for this use case were found by BLAST in nr database using B. subtilis 6S RNA as a query. The search identified 94 unique subject sequences with different level of similarity to B. subtilis 6S RNA and with varying lengths, including fragmented sequences (Text box 3.3).
Text box 3.3. 94 unique sequences including fragments extracted from BLAST search output B. subtilis 6S RNA as a query.
TThe 94 subject sequences from the BLAST output are used as query sequences for cpPredictor to be characterized by the structure of the BLAST query RNA, which is B. subtilis 6S RNA (included in Text box 3.2). To that end, the query sequences and the template structure are copied into the appropriate text boxes in the cpPredictor server submission page (see Figure 3.3).
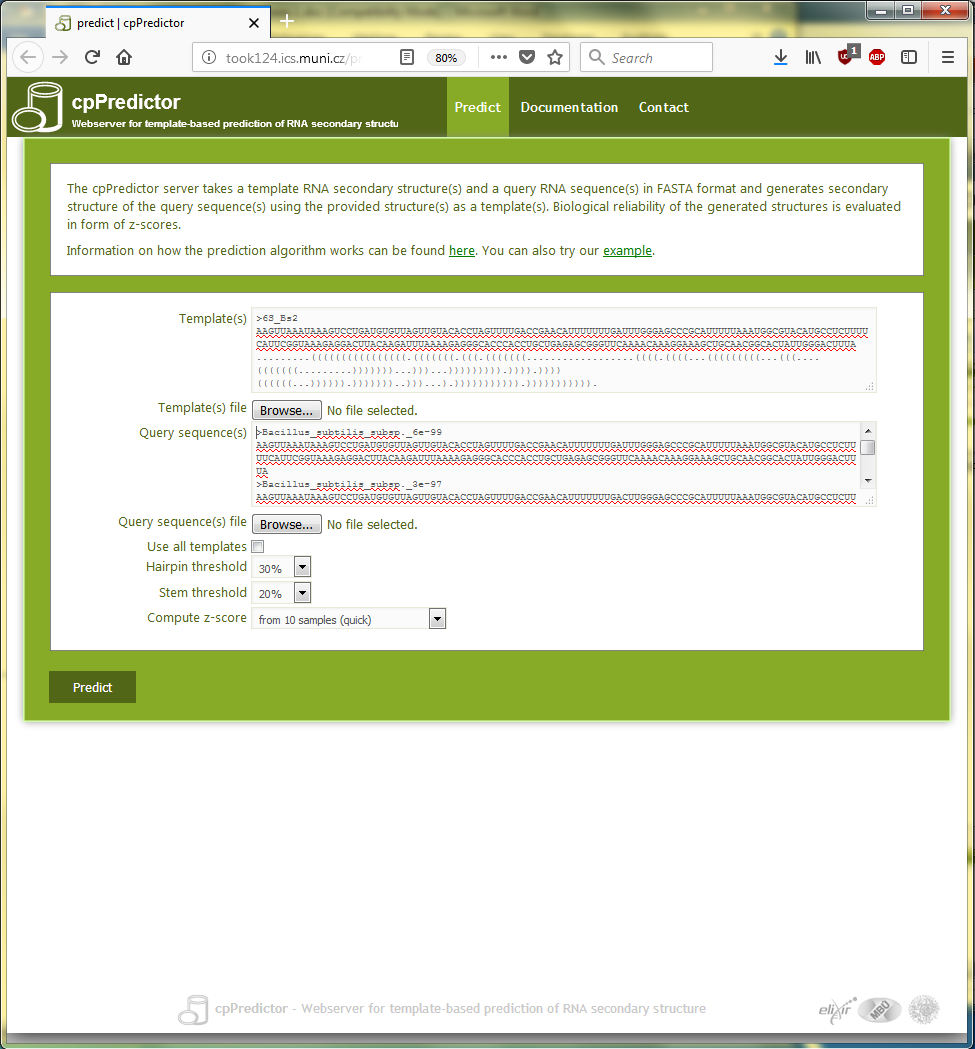
Figure 3.3. The submission page of cpPredictor webserver filled up with query sequences and a template structure for the use case #2: Characterization of RNA sequences including fragments.
cpPredictor generates structures of the 94 sequences. It provides not only secondary structures but also characterizes the sequences with regards to the template structure, including fragments that were also found among the BLAST subject sequences.
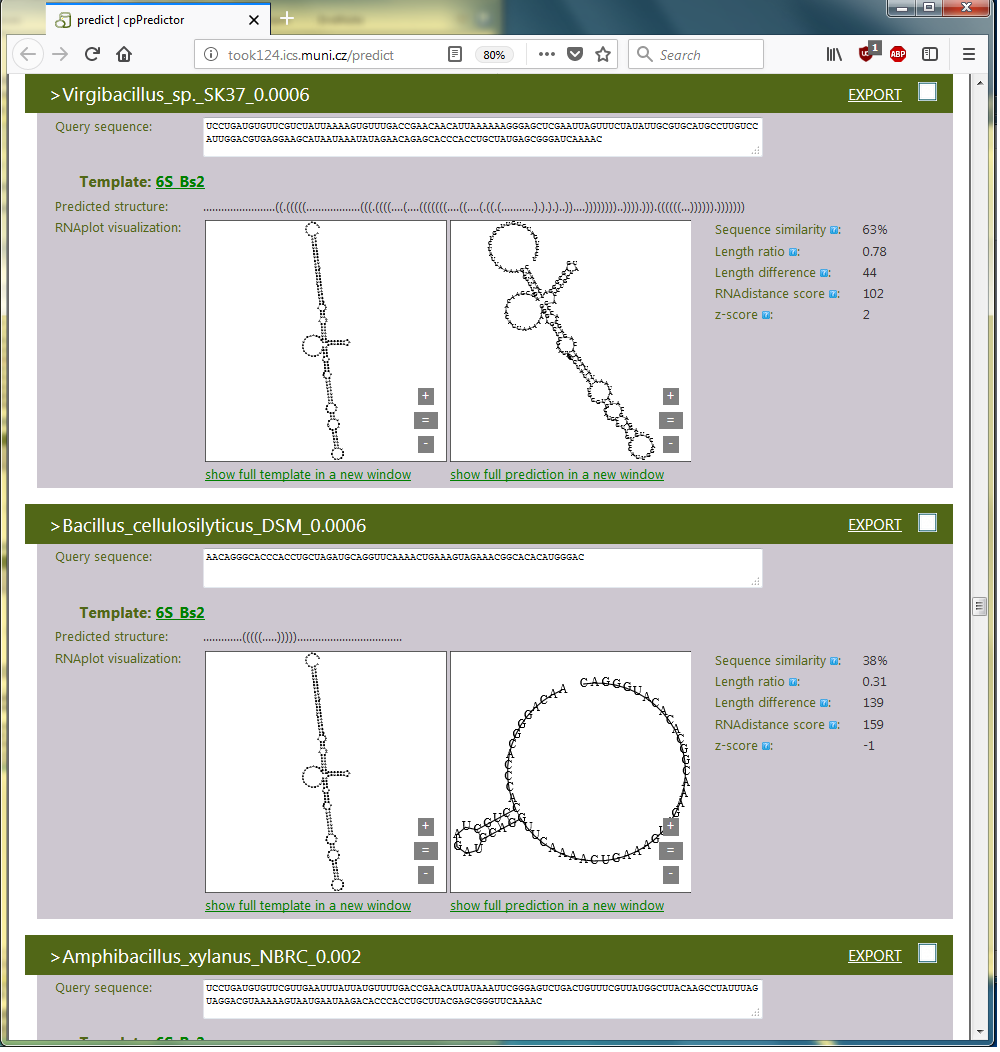
The structural characterization is demonstrated by Figure 3.4 showing two examples of fragments. They were Virgibacillus sp. and B. cellulosilyticus species selected out of the complete cpPredictor output. The first of the fragments is identified as a fragment of 6S RNA by its structure and its z-score. The structure maps the fragment onto the whole 6S RNA structure based on structural similarity. The z-score is ≥ 2, saying that the structure is reliable although it is a fragment, and therefore it most likely represents a substructure of 6S RNA. Indeed, visual comparison to the template maps the substructure to the whole 6S RNA structure. We can not say anything about the other fragment as it is too short.
3.3. Use case #3: Identification of most relevant secondary RNA structure among ambiguous predicted secondary structures¶
This use case demonstrates prediction of RNA secondary structure aided by cpPredictor, when a single RNA has multiple different secondary structures predicted by different algorithms.
For demonstration, H. sapiens u1 RNA is used. Its secondary structure predicted by RNAfold, Mfold and Turbofold differs, i.e. the algorithms produce different structures for the same sequence (see Text box 3.4). The H. sapiens u1 RNA structure predicted by RNAfold, Turbofold and Mfold are denoted u1_hs_C_rf, u1_hs_C_tf and u1_hs_C_mf, respectively.
Text box 3.4. Secondary structure of H. sapiens u1 RNA predicted by RNAfold (u1_hs_C_rf), Turbofold (u1_hs_C_tf) and Mfold (u1_hs_C_mf).
Now the question is, which of the predicted structures is most relevant for H. sapiens u1 RNA. The get the answer, the three predicted structures are used by cpPredictor as templates to generate structures of other homologous u1 RNAs. Here we use 12 RNAs downloaded from Rfam database (Text box 3.5). Their sequences are used as query sequences for each of the templates.
Text box 3.5. Sequences of 12 u1 RNA homologs.
The templates and the query sequences are filled into the cpPredictor input form as shown in Figure 3.5. The check box ‘Use all templates’ checked to predict structure of each query sequence with all 3 templates, which results into 3 structures for each query sequence.
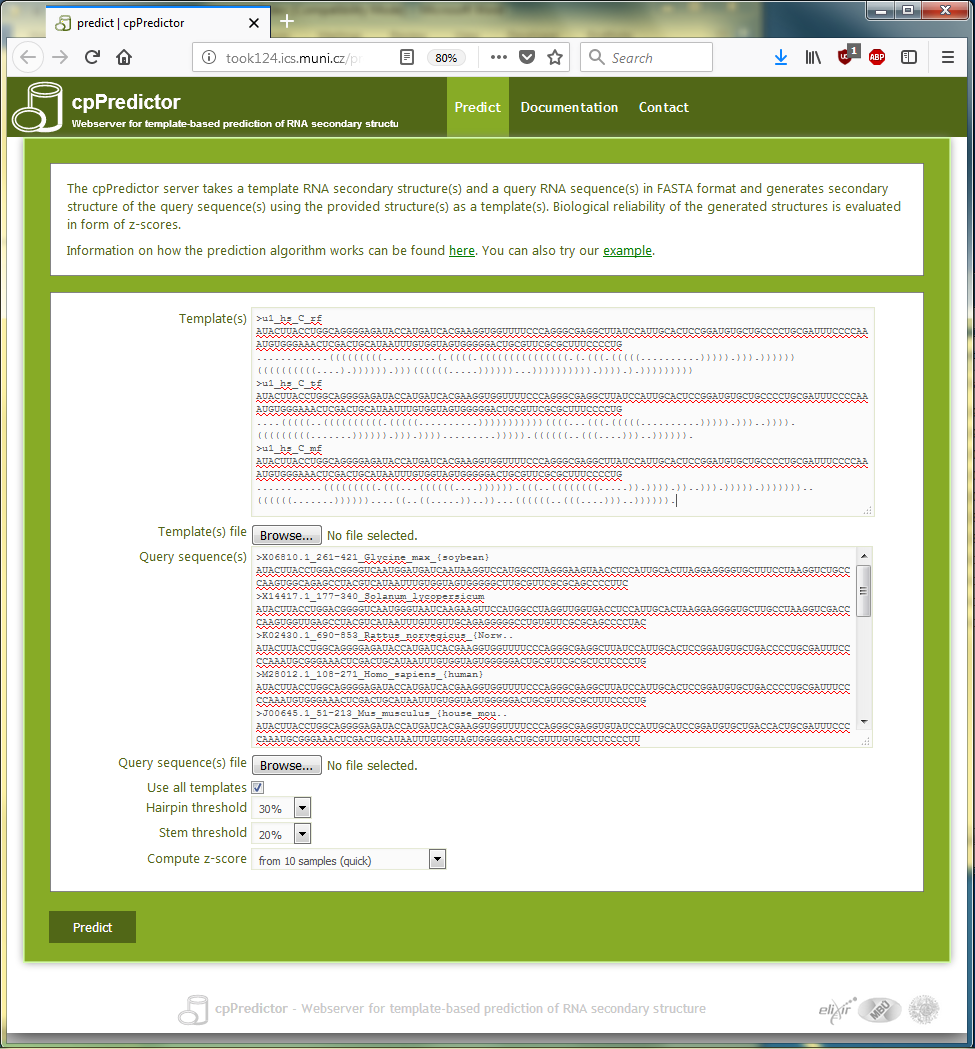
Figure 3.5. The submission page of the cpPredictor webserver filled up with 3 templates and sequences of 12 u1 RNA homologs used as query sequences.
The structures are generated. The templates, as they are different structures, produce 3 different structures for each query sequence (see Text box 3.6).
Text box 3.6. Secondary structures of 12 u1 RNA homologs generated by cpPredictor using 3 templates.
The generated structures are downloaded from the cpPredictor output form. Each of the 3 templates produced 12 structures. The template that is most relevant for u1 RNA structure will generate most consistent structures of the 12 homologs, as its structure most suits the sequences of the homologs. The consistence of the generated structures can be measured using their mutual similarity.
In this use case, we have 3 templates that generated 3 sets of structures. The mutual similarity is computed for structures within the sets by RNAdistance. The highest mutual similarity was found for the set of structures generated using the structure predicted by Turbofold. This template generated most consistent structures of 12 homologous u1 RNAs. The 3 templates and examples of structures generated using them for Solanum lycopersicum u1 RNA are shown in Figure 3.6.
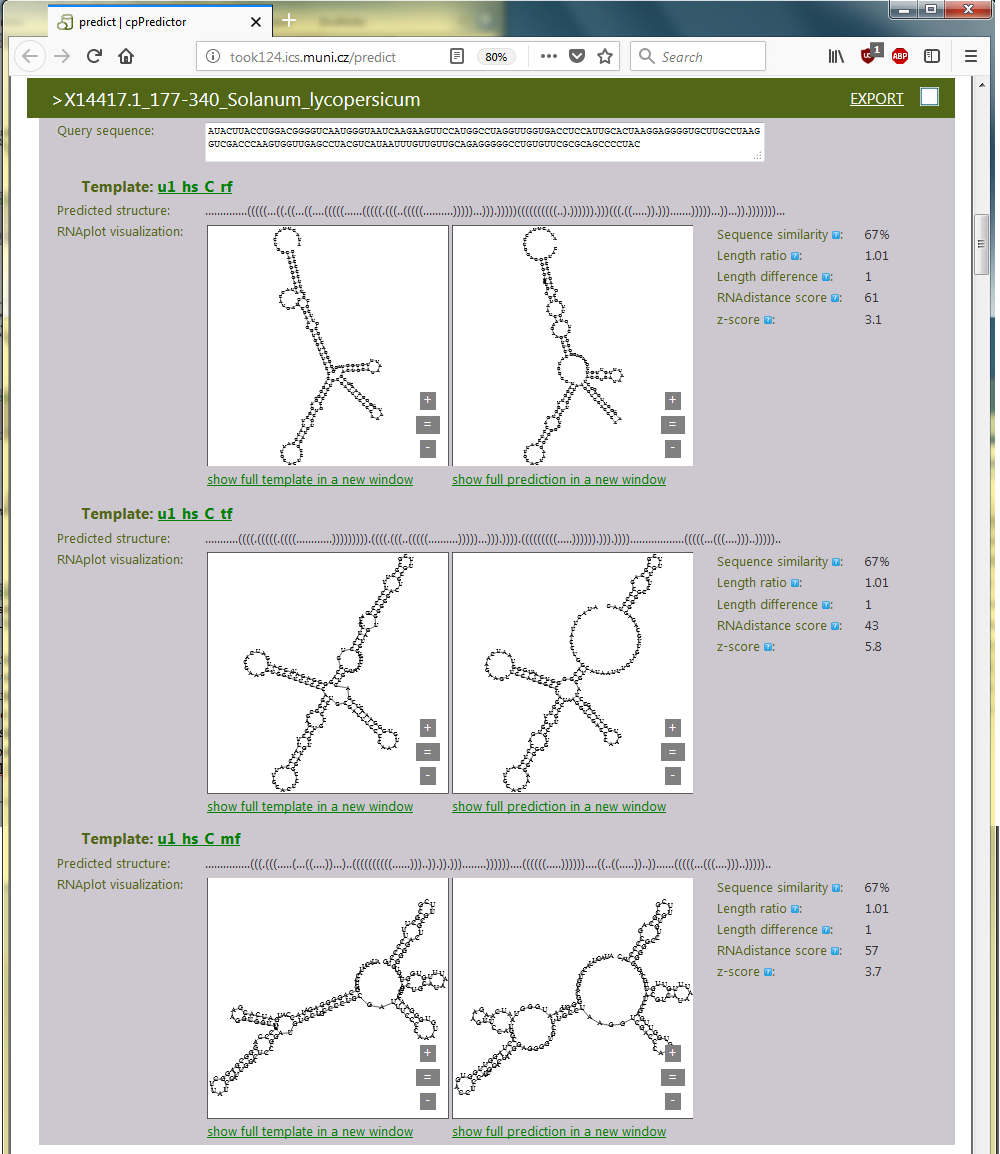
Figure 3.6. An example of generated structures for Solanum lycopersicum u1 RNA using three different predicted structures of H. sapiens u1 RNA as templates.
Indeed, the 2nd template, which is the H. sapiens u1 RNA structure predicted by Turbofold, is similar to the known, physiological H. sapiens u1 RNA structure (cf. Figure 3.6 and Figure 3.7). It is the most similar one among the three predicted structures used as templates, whose structures are included in Figure 3.6. Consequently, the structures generated by cpPredictor using the 2nd template are also similar to the experimentally identified u1 RNA structure, as they were generated by the most relevant template. This is demonstrated in Figure 3.6 for S. lycopersicum u1 RNA.