2. Example¶
The following example demonstrates the basic use of cpPredictor. Specific problems related to RNA secondary structure prediction that can be worked out with the aid of cpPredictor are demonstrated in the section 3 Use cases of this documentation.
The basic aim of cpPredictor is template-based prediction of secondary structure for individual RNA sequences. It is typically the situation, when one has RNAs with sequences but without known structures and wants to get their secondary structures provided that a structural template related to the RNAs is available, and the prediction using available methods is not accurate.
Such an example is ryhB RNA. There are 5 ryhB homologs (Text box 2.1) and experimentally identified structure of one of them, the E. coli ryhB RNA (Text box 2.2), originally described in Masse and Gottesman (2002). The structure is used as a template and the sequences as query sequences for cpPredictor. To fill up the cpPredictor input form with the input data, click the example link on the cpPredictor server input page. Note, that the structures of the ryhB homologs can not be predicted accurately with available tools including RNAfold (also in a constrained mode using the known structure as a constraint), Mfold and Turbofold, except for E. coli ryhB RNA, whose secondary structure is predicted correctly by Mfold.
Text box 2.1. Sequences of 5 ryhB RNAs.
Text box 2.2. Structure of E. coli ryhB RNA.
The query sequences in Text box 2.1 and the template structure in Text box 2.2 are copied into the appropriate text boxes on the submission page (shown in Figure 2.1. To get z-scores for the predicted structures, the ‘Compute z-scores from 10 samples (quick)’ choice in ‘Compute z-score’ item is selected.
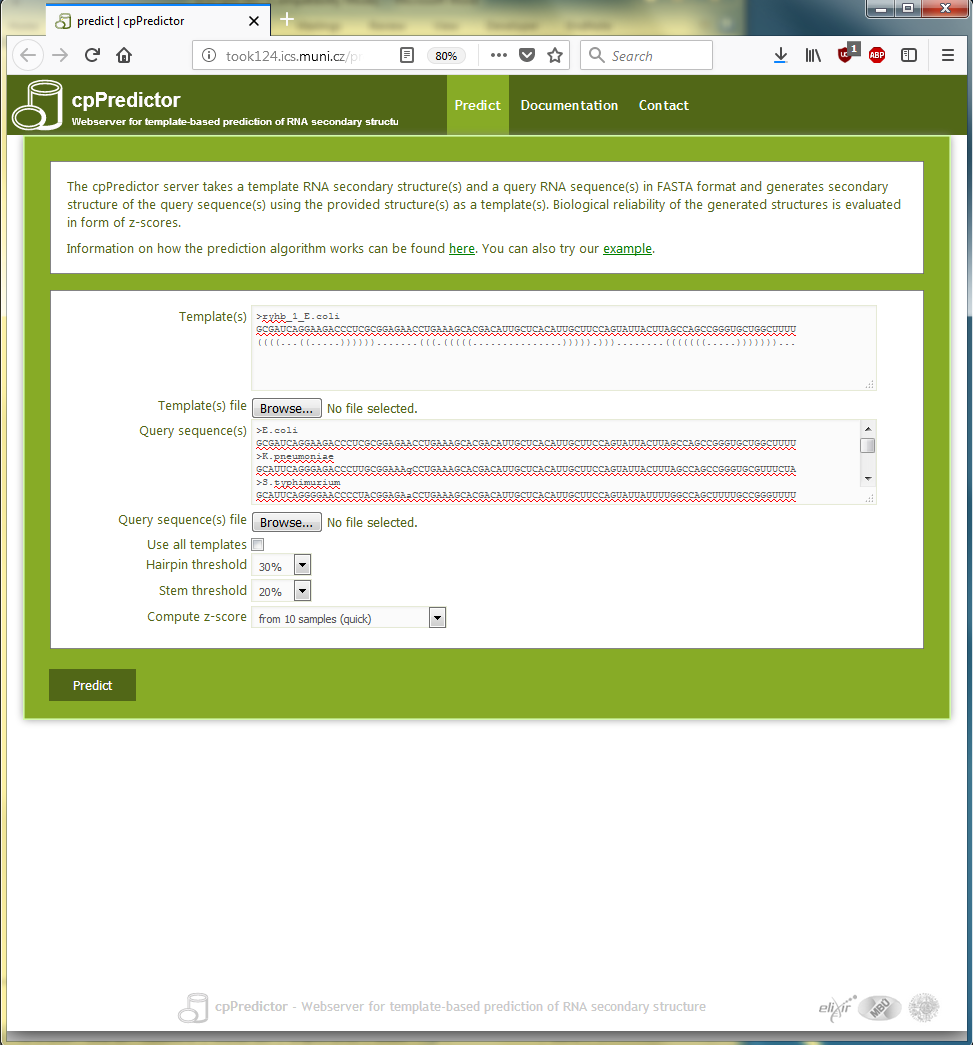
Figure 2.1. The submission page of cpPredictor web server filled up with query sequences and a template structure for the example.
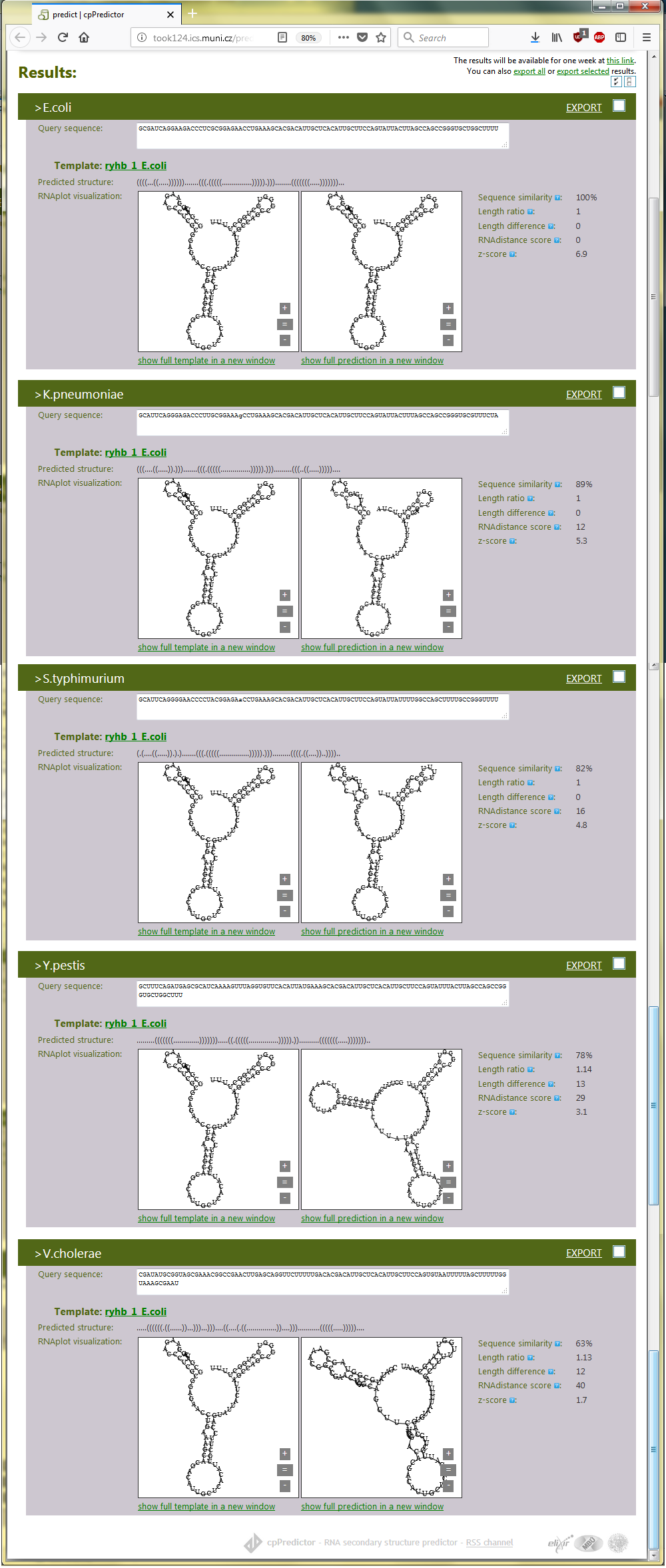
The cpPredictor output consists of sections for each of query RNAs. The sections consist of template structure (the left panel) and predicted structures (the right panel) together with other sequence and structure characteristics of query RNAs. The items in the output form are accompanied with quick help indicated by question marks, making their use and function clear. The template structure and the generated structures are placed side by side so they can be compared visually. For the example of ryhB RNAs, the output is shown in Figure 2.2.
cpPredictor generated secondary structures of ryhB homologs and their z-scores. Except for V. cholera ryhB RNA, the generated structures have z-score > 2 indicating their reliability. z-score of V. cholera ryhB RNA is < 2, although is close to 2, owing to its larger evolutionary distance from E. coli, that is indicated by relatively low sequence similarity to the template sequence, 63%. The similarity is also included in the cpPredictor output page.
Note, that the computation is quick; cpPredictor is aimed at fast and robust characterization of RNA sequences by their secondary structures. The predicted secondary structures can be exported via check boxes placed above on the right in the title bar of the output sections.